The CRISPR/Cas9 system is a revolutionary genome editing tool. However, in eukaryotes, search and optimization of a suitable promoter for guide RNA expression is a significant technical challenge.
Recently, the research group led by Prof. SUN Jibin and Prof. ZHENG Ping used the industrially important fungus, Aspergillus niger, to demonstrate that the 5S rRNA gene, which is both highly conserved and efficiently expressed in eukaryotes, can be used as a guide RNA promoter. This novel sgRNA-expressing strategy based on 5S rRNA internal promoter was able to significantly improve the editing efficiency of the CRISPR/Cas9 system in A. niger. This system enabled precise edits in a single gene locus or multiplex gene loci and removed 48 kb large genomic region containing contiguous gene clusters necessary for mycotoxin biosynthesis from the genome in the demonstration. Based on this discovery, they established a powerful and highly efficient genome editing toolbox for A. niger, which will dramatically promote the fundamental understanding of the peculiarities of A. niger and accelerate its biotechnological application as a universal cell factory for better performance or new products. Moreover, this sgRNA expression strategy based on 5S rRNA might constitute a novel and universal solution to build expression cassettes for sgRNA, which will accelerate the implementation of CRISPR/Cas9 system in other eukaryotic organisms.
The study entitled “5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing inAspergillus niger” has been published in ACS Synthetic Biology . Dr. ZHENG Xiaomei, assistant researcher of TIB, is the first author of this paper. This study was supported by the National Natural Science Foundation of China (Grant No, 31700085, 31370113 and 31370829) and partially by the Chinese Academy of Sciences President’s International Fellowship Initiative CAS PIFI (Grant No, 2018VBA0013 and 2018PB0036).

Figure 1. 5S rRNA based sgRNA expression cassette enables the CRISPR/Cas9 system to disrupt the albA gene in A. niger.
(A) Schematic diagram of albA disruption mediated by NHEJ using CRISPR/Cas9 based on U6 and 5S rRNA promoters. (B) The prediction of different genetic elements of the 5S rRNA gene and the six-truncation setup. (C) albA gene disruption efficiency of the transformants with sgRNA constructs driven by different promoters.
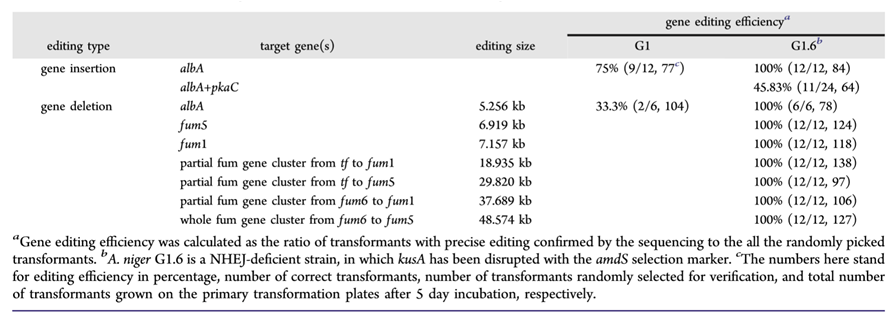
Table 1. Genome precise editing efficiency based on 40-bp homologous arms achieved in this study
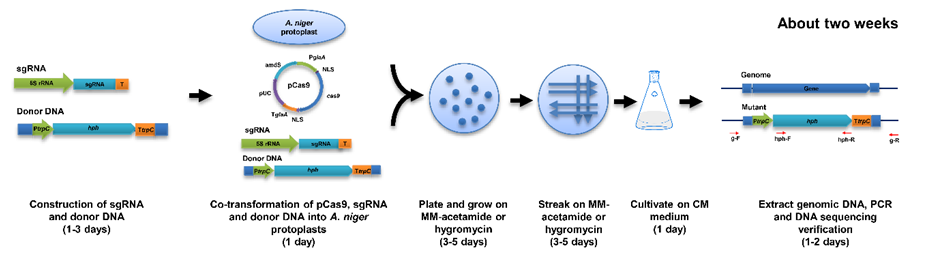
Figure 2. Standard protocol for the novel 5S rRNA-based CRISPR/Cas9 system for A. niger
Contact:Prof. ZHENG Ping
Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences.
Email: zheng_p@tib.cas.cn