With the expansion of fruit plants at the end of the Cretaceous period, the ancestor of the Saccharomyces lineage generated a novel trait of aerobic fermentation to survive in an environment with abundant fermentable fruits. During aerobic fermentation, the yeast can repress the expression of mitochondrial genes, rapidly consume glucose, accumulate ethanol, and out-compete other microbes. Therefore, the formation mechanism of aerobic fermentation is always a hotspot.
Many studies have shown that the whole genome duplication (WGD) event that happened around 100 million years ago played an important role in the origin of aerobic fermentation in Saccharomyces. Our study found the other wine and beer yeast Dekkera bruxellensis diverged from the Saccharomyces cerevisiae about 300 million years ago, long before WGD, also generated the capacity of aerobic fermentation. By profiling and comparing genome sequences, transcriptomic landscapes and chromatin structures, we revealed the fact that no WGD occurred in Dekkera lineage. Surprisingly, the two independent evolutionary lineages evolved similar AT-rich elements in promoter regions of mitochondrial genes, which underlay the parallel changes in chromatin structure and led to concerted suppression of mitochondrial functions by glucose and metabolic convergence in these two independent yeast species. The study confirmed that similar genetic mechanism and metabolic pathway can emerge in independent evolutionary lineages for improving adaptability in the same conditions.
The study entitled “Parallel Evolution of Chromatin Structure Underlying Metabolic Adaptation” has been published in Molecular Biology and Evolution. Dr. CHENG Jian, assistant professor of TIB and Dr. GUO Xiaoxian , Postdoc of Cornell University, are co-first authors of this paper. Prof. JIANG Huifeng and Prof. GU Zhenglong are co-corresponding authors of this paper. This research was supported by two NSFC grants (Grant No, 31300077 and 31470215) and NSF grant MCB-1243588 to Z.G. 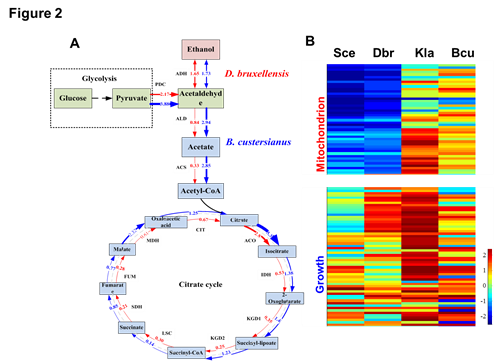
Parallel evolution of gene expression between D. bruxellensis and S. cerevisiae. (A) Relative gene expression for genes from catabolic pathways and the TCA cycle. (B) Relative gene expression for mitochondrial genes and growth associated genes in YPD compared to YPEG.(Image by TIB) Contact: Prof. Huifeng Jiang
Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences. Email: jiang_hf@tib.cas.cn |