Lysine acetylation is an important type of reversible post-translational modification (PTM) that takes place in the ε-amino group of lysine residues in proteins by an enzyme-catalyzed chemical reaction that adds an acetyl group to an organic compound. It plays an important role in myriad biological processes, including cell apoptosis, DNA-protein interaction, DNA replication and repair, DNA transcription and protein stability. Despite its importance, our understanding of the regulatory roles of lysine acetylation remains nebulous. Identification of acetylation sites is an essential first step towards elucidation of the mechanism underlying protein acetylation.
The Laboratory of Structural Bioinformatics and Integrative System Biology led by Dr. SONG Jiangning at Tianjin Institute of Industrial Biotechnology (TIB), Chinese Academy of Sciences (CAS) carried out a series of in-depth analysis of species-specific lysine acetylation, in close collaboration with researchers from the Bioinformatics Center, China Agricultural University and the Faculties of Medicine and Information Technology at Monash University, Melbourne Australia. The researchers have recently developed a powerful bioinformatics tool termed as SSPKA (http://www.structbioinfor.org/Lab/SSPKA/) (See Figure 1 for a screenshot of the interface and example output of the SSPKA tool) for species-specific lysine acetylation prediction. This tool combines sequence-derived and functional features with two-step feature selection to build the models. Feature importance analysis indicates functional features, applied for lysine acetylation site prediction for the first time, significantly improve the predictive performance.
Then the researchers apply the SSPKA model to screen the entire human proteome and identify many high-confidence putative substrates that are not previously identified. The results along with the implemented Java tool, serve as useful resources to elucidate the mechanism of lysine acetylation and facilitate hypothesis-driven experimental design and validation.
This bioinformatics tool represents a new alternative approach to study the complicated relationship between sequence, structure and function of acetylated proteins.
This work was supported by grants from the Knowledge Innovation Program of CAS (KSCX2-EW-G-8) and National Natural Science Foundation of China (61202167, 61303169 and 11250110508). The paper entitled “Accurate in silico identification of species-specific acetylation sites by integrating protein sequence-derived and functional features” has been published in the July Issue of Scientific Reports (http://www.nature.com/srep/2014/140721/srep05765/full/srep05765.html). Mr. LI Yuan, a recent Master graduate student from TIB, is the first author of this paper.
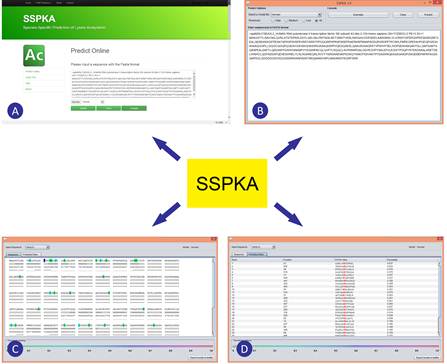
Screenshot of the interface and example output of the SSPKA tool. (A) Screenshot of the web server; (B) Screenshot of sequence submission of the local Java applet; (C) and (D) Screenshot of prediction output of the local Java applet. (Image by SONG Jiangning’s group)