The MSE (where MSE is low energy (MS) and elevated energy (E) mode of acquisition) acquisition method commercialized by Waters on its Q-TOF instruments is regarded as a unique data-independent fragmentation approach that improves the accuracy and dynamic range of label-free proteomic quantitation. Due to its special format, MSE acquisition files cannot be independently analyzed with most widely used open-source proteomic software specialized for processing data-dependent acquisition files.
In this study, the researchers, who from the Laboratory led by Professor SHUI Wenqing at Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, established a workflow integrating Skyline, a popular and versatile peptide-centric quantitation program, and a statistical tool DiffProt to fulfill MSE-based proteomic quantitation. Comparison with the vendor software package for analyzing targeted phosphopeptides and global proteomic datasets reveals distinct advantages of Skyline in MSE data mining, including sensitive peak detection, flexible peptide filtering, and transparent step-by-step workflow.
Moreover, researchers developed a new procedure such that Skyline MS1 filtering was extended to small molecule quantitation for the first time. This new utility of Skyline was examined in a protein–ligand interaction experiment to identify multiple chemical compounds specifically bound to NDM-1 (where NDM is New Delhimetallo-β-lactamase 1), an antibiotics-resistance target. Further improvement of the current weaknesses in Skyline MS1 filtering is expected to enhance the reliability of this powerful program in full scan-based quantitation of both peptides and small molecules.
The study entitled “Exploring skyline for both MSE-based label-free proteomics and HRMS quantitation of small molecules” has been published in Proteomics (2014,14: 169–180). LIU Shanshan (a graduate student trained in TIB and Nankai University) is the first author of this paper. This work was supported by the National Natural Science Foundation of China (31170782) and Tianjin Natural Science Foundation (11JCYBJC25500).
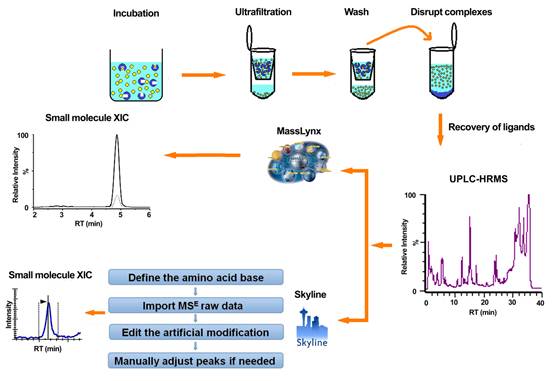
MSE-based label-free proteomic quantitation workflow(Image by SHUI Wenqing's group)