Pectate lyase (Pel, EC 4.2.2.2) is capable of cleaving α-1,4 linkages in pectate polymers by β-elimination mechanism, producing 4,5-unsaturated oligogalacturonates, and has attracted a great deal of interest in the past few decades from both scientific and commercial perspectives. Biotechnological applications of microbial pectate lyases (Pels) in plant fiber processing are considered as environmentally friendly. As such, they become promising substitutes for conventional chemical degumming process. Since applications of Pels in various fields are widening, it is necessary to explore new pectolytic microorganisms and enzymes for efficient and effective usage.
Pectolytic enzymes are ubiquitous in nature, and most of the Pels previously reported have been isolated from microorganisms such as bacteria and fungi. Industrial applications of bacterial alkaline Pels, the majority of which occur in the Bacillus and Pseudomonas genus, have been extensively studied. However, in terms of practical bioprocesses, only a few of them are robust enough to undergo long-time bioprocess treatment. Recently, enzymes originating from thermophilic or alkaliphilic microorganisms are garnering more interest because of their good performance at extreme temperature or pH. Although only limited information on the alkaliphilic strains of Paenibacillus spp. exists, several Pels from this genus have proved to be highly stable. Hence, these bacteria have great potential for industrial-scale production of novel enzymes with desirable compatibility between the optimal temperature/pH and the extended useful life.
In this study, a Pel gene (pelN) was cloned using degenerate PCR and inverse PCR from the chromosomal DNA of Paenibacillus sp. 0602 by the Laboratory of Structural Bioinformatics and Integrative System Biology led by Professor SONG Jiangning at Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences. This enzyme belongs to the polysaccharide lyase family 1. The purified recombinant enzyme exhibited a specific activity of 2,060 U/ mg on polygalacturonic acid, and the maximum Pel activity produced by Escherichia coli in shake flasks reached 2,467.4 U/mL, which represent the highest level in shake-flask fermentation of Pel. The maximum activity was observed in a buffer with 5 mM Ca2+ at pH 9.8 and 65°C. PelN displays a half-life of around 9 h and 42 h at 50°C and 45°C, respectively. The biochemical treatment achieved the maximal reduction of percentage weight (30.5%) of the ramie bast fiber.
This work represents the first study that describes the extracellular expression of a Pel gene from Paenibacillus species in E. coli. The high yield of the extracellular overexpression, relevant thermostability and efficient degumming using combined treatments indicate its strong potential for large-scale industrial production.
The paper entitled “Cloning, recombinant expression and characterization of a pectate lyase from Paenibacillus sp. 0602 in recombinant Escherichia coli” has been published in BMC Biotechnology. LI Xiaoman and WANG Huilin, currently Research Assistants in this lab, shared the first authorship of this paper. In addition, two Chinese patents were also submitted (201210064549.8 and 201310155628.4). This research was supported by grants from the Knowledge Innovation Program of CAS (KSCX2-EW-G-8), Tianjin Municipal Science & Technology Commission (10ZCKFSY05600).
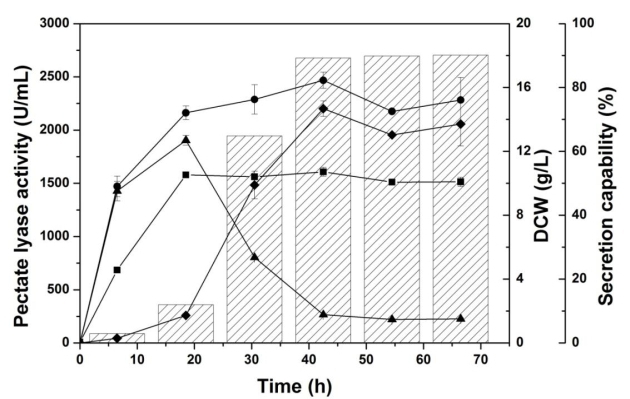
Time profiles for shake-flask cultivation in 250-mL flasks. Intracellular Pel activity (▲), extracellular Pel activity (◆), total Pel activity (●), cell concentration (■), and secretion capability (sparse column).
(Image from Prof. SONG Jiangning’s group)