Caspases and granzyme B (GrB) are important proteases involved in fundamental cellular processes and play essential roles in programmed cell death, necrosis and inflammation. Although a number of substrates for both types have been experimentally identified, the complete repertoire of caspases and granzyme B substrates remained to be fully characterized. Accordingly, systematic bioinformatics studies of known cleavage sites may provide important insights into their substrate specificity and facilitate the discovery of novel substrates. We develop a new bioinformatics tool, termed Cascleave 2.0 (http://www.structbioinfor.org/cascleave2/), which builds on previous success of the Cascleave tool for predicting generic caspase cleavage sites. It can be efficiently used to predict potential caspase-specific cleavage sites for the human caspase-1, 3, 6, 7, 8 and GrB. In particular, we integrate heterogeneous sequence and protein functional information from various sources to improve the prediction accuracy of Cascleave 2.0. During classification, we use both maximum relevance minimum redundancy and forward feature selection techniques to quantify the relative contribution of each feature to prediction and thus remove redundant as well as irrelevant features. A systematic evaluation of Cascleave 2.0 using the benchmark data and comparison with other state-of-the-art tools using independent test data indicate that Cascleave 2.0 outperforms other tools on protease-specific cleavage site prediction of caspase-1, 3, 6, 7 and GrB. Cascleave 2.0 is anticipated to be used as a powerful tool for identifying novel substrates and cleavage sites of caspases and GrB and help understand the functional roles of these important proteases in human proteolytic cascades. 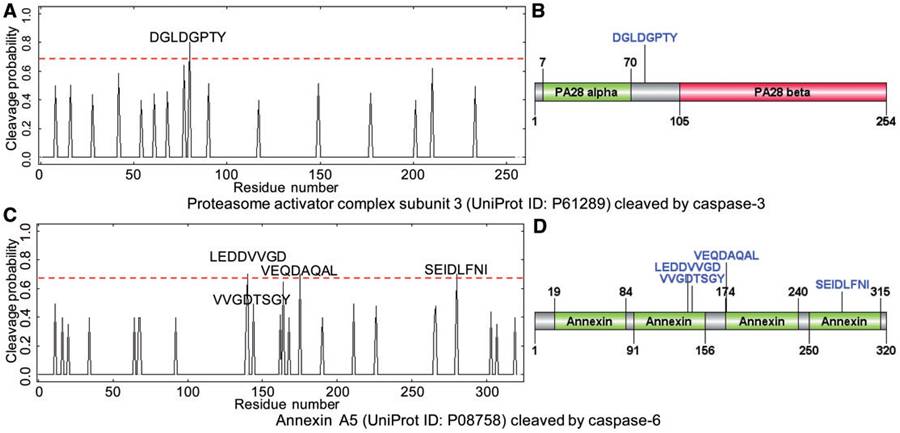
The predicted cleavage probability for caspase cleavage sites using Cascleave 2.0. (A) Sequence-scanning results and (B) predicted cleavage site locations with respect to the domain of proteasome activator complex subunit 3; (C) sequence-scanning results and (D) predicted cleavage sites with respect to the protein domain of Annexin A5 (Uniprot ID: P08758). |