CRISPR genome editing techniques employing deaminases fused to Cas9 variants were developed for precise genomic base editing (BE). However, those BEs can yield only base transitions, including C-to-T (or G-to-A) and A-to-G (or T-to-C) substitutions; they cannot produce base transversions. To fill these technological gaps, ZHANG Xueli and BI Changhao groups developed new base editors named as glycosylase base editors (GBE), as shown in Fig. 1, an enzyme complexes of fused nCas9, AID, and Ung, these fusions perform a series of functions, including specific DNA binding, cleaving the amine group from C, and excising U to create AP sites followed by cellular repair system, and achieve specific base editing.
In wild-type Escherichia coli strains, GBE approach was found to convert C to A with an average specificity of 93.8±4.8%, an any base editing (NBE) was also developed, where, in a one, two or three-step process, any A, T, G or C could be converted to any other base (Fig. 2). In mammalian cells, GBE achieved C-to-G conversions with a high specificity at the 6th C in an N20 sequence, which is different from other BE techniques. Out of thirty tested loci, two loci had a C-to-G specificity of more than 90%, seven loci were 80% to 90%, eight loci were 70% to 80%, six loci were 50% to 70%.
The GBE technique is the first base editing technique that allows specific C-to-A conversion and any base editing in bacteria, and also the first C-to-G conversion in mammalian cells, with high G/C conversion specificity and position specificity, GBE could be used as a major genome editing technique. As a new generation of base editing technology, GBE directly modifies the target base instead of relying on DNA replication. This technology further improves the base editing system, fills in the vacancy of the existing base editing technology, and realizes the arbitrary base editing of microbes for the first time, which greatly improves the ability gene editing and synthetic biotechnologies. GBE is also the first base editor that can perform C-G specific transversion in mammalian cells. With high specificity and narrow editing window, GBE will bring light to the treatment of human genetic diseases caused by more than 3000 known CG base mutations (Fig. 3). The breakthrough of this base editing technology has further improved the basic capability of China's biotechnology, and will play an important role in the independent innovation of the core key technologies of the biological industry.
The study entitled “New base editors change C to A in bacteria and C to G in mammalian cells” has been published in Nature Biotechnology. ZHAO Dongdong, a research assistant of TIB, LI Ju, a professor of Tianjin Normal University and LI Siwei, a research assistant of TIB are the co-first author of this paper. BI Changhao and ZHANG Xueli, professors of TIB, are the corresponding authors of this paper. A PCT patent was also applied. This research was supported by the National Key Research and Development Program of China, the Key Research Program of the Chinese Academy of Science, National Natural Science Foundation of China and Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project.
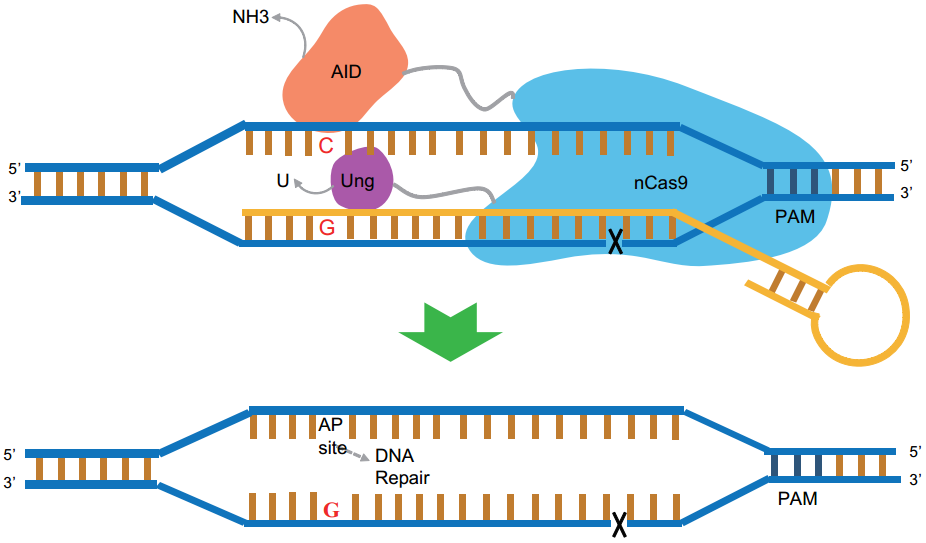
Figure 1. Illustration of the functional mechanism (Image by Prof. BI Changhao’s group)
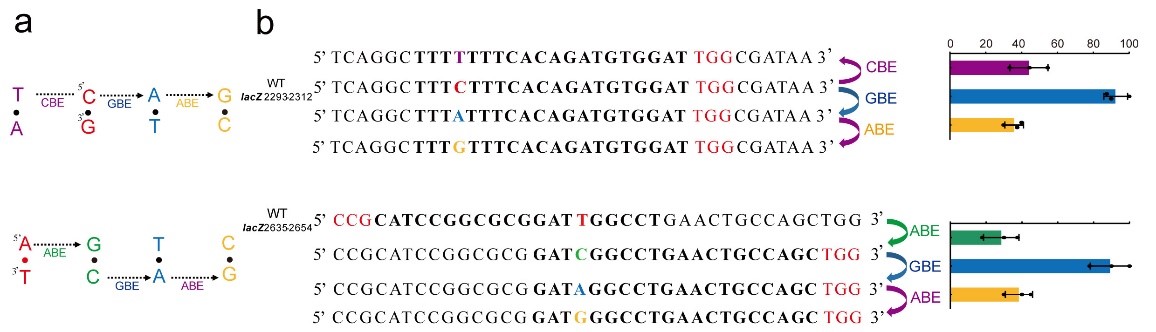
Figure 2. Any base editing (NBE) in E. coli (Image by Prof. BI Changhao’s group)
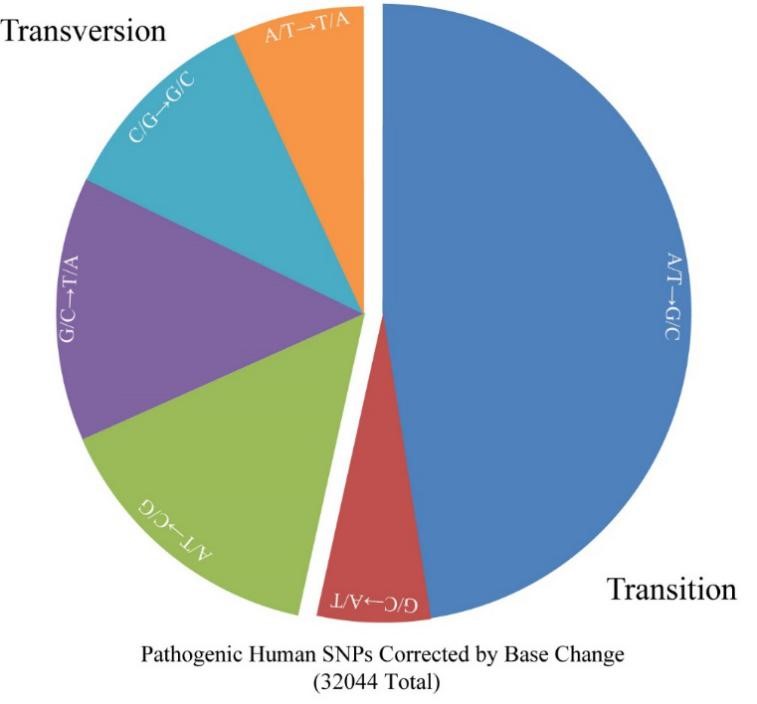
Figure 3. Base pair changes required to correct pathogenic human SNPs (Image by Prof. BI Changhao’s group)
Contact:Prof. BI Changhao
Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences.
Email: bi_ch@tib.cas.cn