The conversion ratio from substrates to target products is one of the important objectives for industrial strains and is also a main cost control in the fermentation process. Knocking out specific genes or specific pathways is one of the main means to achieve this goal by reducing the entailed loss of byproduct synthesis and enhancing the backbone fluxes. Currently, some algorithms for knockout target predictions, such as OptKnock, RobustKnock and GDLS, utilized mixed integer bi-level linear programming (MIBLP) to coordinate cellular growth and bioengineering objective, but these algorithms can only give single knockout strategy while solving takes long time.
Based on cell metabolic networks, multi-strategy prediction method, named ReacKnock, for enzyme knockouts was developed by the computational biologists from the Laboratory led by Professor SUN Jibin at Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences. KKT (Karush-Kuhn-Tucker) technology, which was strictly verified in mathematics, was used to solve mixed integer bi-level linear programming and KKT can guarantee the correctness of the prediction results. The solving time was greatly shortened for the concision of ReacKnock and especially it can provide multi-knockout strategies.
Based on the genome scale metabolic network of E.coli, as for 9 typical chemicals including Succinate, Ethanol, Acetate, Hydrogen, Formate, Glycolate, Lactate, Fumarate and Threonine, both ReacKnock and OptKnock were carried out for prediction and comparison. The computational results on virtual mutant strains were verified by FVA (Flux Variability Analysis) for the maximum production and by FBA (Flux Balance Analysis) for the maximum growth. Results showed that ReacKnock would provide knockout predicts with higher conversion ratios from substrates to target products, as shown in Table 1; Due to the high-precision on solution, the results of ReacKnock were fully in line with the FVA verification for production and the FBA verification for growth; ReacKnock could provide all the alternative strategies in the same deletion number, Table 2 illustrating the first 10 knockout strategies for 6 enzyme composition as for succinic acid production with E. coli; To computing speed, because of the concision in algorithm, ReacKnock was faster.
The method from this study can provide all the alternative knockout strategies in the same deletion number, can provide the design of genetic engineering with more ideas and choices for industrial strain development, and is advantageous for the experiment. The method has universality, applicable to all industrial microorganisms, can be calculated on the genome scale metabolic network and then guide the experiments.
The paper entitled "ReacKnock: identifying reaction deletion strategies for microbial strain optimization based on genome-scale metabolic network" has been published in PLoS ONE. XU Zixiang, a researcher on computational biology in TIB, is the first author of this paper. This research was supported by grants from National Basic Research Program of China, National High Technology Program, National Natural Science Foundation of China and Key Technologies Research and Development Program of Tianjin.
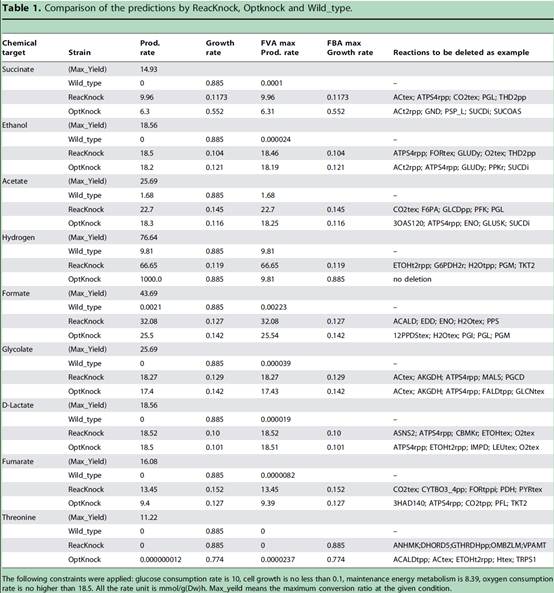
Tablet 1. Knockout prediction and validation of ReacKnock, to improve conversion ratios of 9 chemicals with E. coli
(Image by SUN Jibin's group)
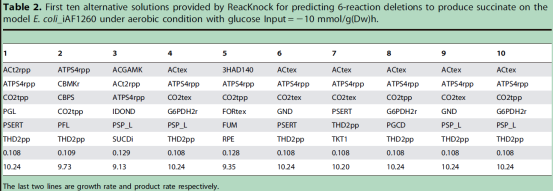
Table 2. 10 alternative strategies provided by ReacKnock for the production of succinic acid with E.coli
(Image by SUN Jibin's group)