July 16, 2013 ------ Dr. Huifeng Jiang’s lab published an article on PNAS with the title “High-resolution view of bacteriophage lambda gene expression by ribosome profiling”. 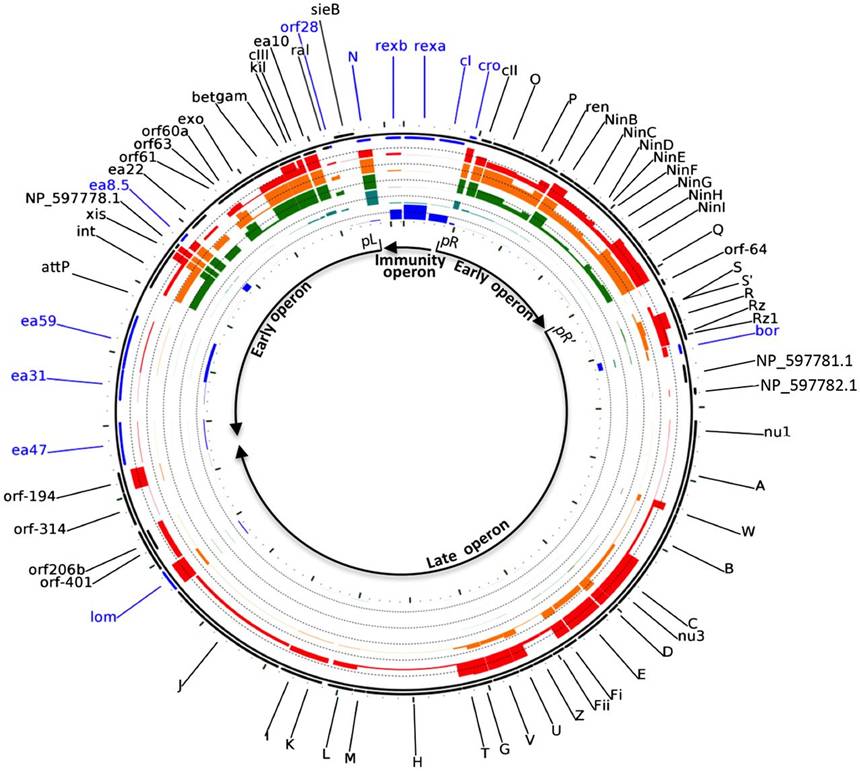
Bacteriophage lambda is one of the most extensively studied organisms and has been a primary model for understanding basic modes of genetic regulation. Both the classic, focused analysis of gene expression and the modern global approaches such as array hybridization have been applied to elucidate the complex pattern of gene function during lambda growth. A recent method, ribosome profiling, provides a further detailed and precise view of gene expression by capturing the instantaneous translation sites of all of the ribosome in a cell. In this work, the authors applied ribosome profiling to the process of lytic growth of bacteriophage lambda to map in detail the expression of lambda proteins and to infer unique loci of translation. They confirmed the known patterns of gene expression, but also show an unexpected complexity of the translation landscape, they found that much ribosome occupancy occurs in ORFs of unknown function. The authors further discovered that ribosome binding can change the structure and availability of mRNA for translation of other parts of the message, or for other functions of RNA such as transcription termination. They also gave another possibility, functional ORFs might be retained in regions of the genome where transcription occurs, even if no functional polypeptide is produced. Thus, they provided the possibility that some or all of these ORFs have no function at all, representing just background activity of the transcription and translation systems. Dr. Huifeng Jiang is one of co-first authors of the paper., and Jeffrey W. Roberts was the corresponding author. 
PNAS, 2013,110(29), pp 11928-11933, DOI: 10.1073/pnas. 1309739110. High-resolution view of bacteriophage lambda gene expression by ribosome profiling. Xiaoqiu Liu, Huifeng Jiang, Zhenglong Gu, and Jeffrey W. Roberts. |